Molecular Structure Determination and Imaging
John D. Furber
Master of Science, Biological Sciences, University of California, Irvine.
Bachelor of Arts, Physics and Mathematics, University of California, Santa Cruz.[ John D. Furber home page ]

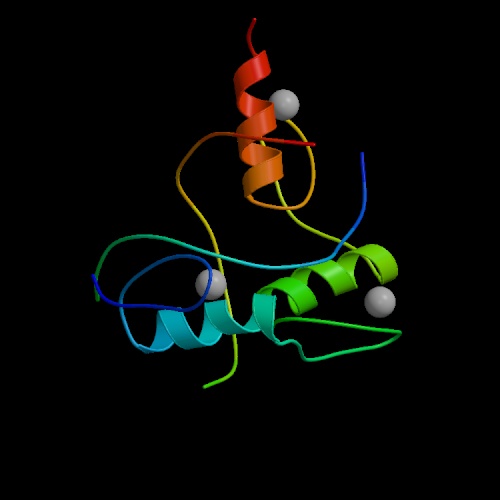 The 3-dimensional structures of bio-molecules can give insight into their mechanisms of function, and can suggest strategies for drug design. A number of bio-molecular structures have been solved by two physical techniques: X-ray Scattering from crystals of purified molecules and Nuclear Magnetic Resonance analysis of molecules in solution.
The 3-dimensional structures of bio-molecules can give insight into their mechanisms of function, and can suggest strategies for drug design. A number of bio-molecular structures have been solved by two physical techniques: X-ray Scattering from crystals of purified molecules and Nuclear Magnetic Resonance analysis of molecules in solution.
After a structure has been solved, then 3D computer models can represent the structure on screen in various forms and from any viewing angle. However, difficulties with the physical methods (X-ray and NMR) have left the structures of many macro-molecules unsolved to date. Work is proceeding slowly on the very difficult problem of computationally predicting accurate 3D structural models solely from sequence data.
The image at left is a model of the "zinc finger" domain of a "zinc finger protein". This class of proteins is able to identify and bind to specific DNA sequences, thereby controlling the expression of specific genes.
- PDB - the Protein Data Bank is an international repository for the processing and distribution of 3-D structure data of biological macromolecules (mainly proteins) determined experimentally by NMR and X-ray crystallography. Until recently, the PDB was maintained by the Brookhaven National Laboratory in New York. However, a new nonprofit organization, the Research Collaboratory for Structural Bioinformatics (RCSB), is now operating the Protein Data Bank under contract to the U.S. Government. http://www.rcsb.org/pdb/index.html
- Protein Explorer and RasMol are free software utilities (by Roger Sayle of Glaxo Wellcome) for imaging molecules on your computer screen. They can visualize proteins, nucleic acids, and small molecules. They can use the coordinates downloaded from PDB. Interactive controls allow rotating molecules and highlighting features. Images can then be saved as GIF files, and scripts can be saved for viewing in Chime. Available from the University of Massachusetts RasMol home page: http://www.umass.edu/microbio/rasmol/
- Chime is a free web browser plug-in by MDL Information Systems, Inc. for viewing molecules if they have been embedded in a web page by its author. Authors and educators can prepare scripted animations with Chime, and place them in web pages. Additional information and tutorials are available from the University of Massachusetts RasMol home page: http://www.umass.edu/microbio/rasmol/ and from MDL Information Systems, Inc. http://www.mdli.com
- Kinemage is free software (by David C. Richardson of Duke University) for interactively viewing molecular structures on your computer screen using the coordinates downloaded from PDB. Interactive controls allow zooming and rotating whole molecules as well as rotating parts of molecules about sigma bonds. http://kinemage.biochem.duke.edu/website/kinhome.htm
- The Online Macromolecular Museum produced at Kenyon College guides you through tutorials with Chime-animated molecules.
- Introduction to Protein Structure (Second Edition, Garland Publishing) is an excellent textbook by Carl Branden and John Tooze.
- The Argonne National Laboratory Structural Biology Center. An Argonne press release describes their work at http://www.anl.gov/OPA/frontiers/b1excell.html.
- A map or "Periodic Table" of protein motif structures is being organized at UC Berkeley. The work is detailed in PNAS, Feb 2003, and described in a news story on Bio.com.
- The Indiana University Molecular Structure Center http://www.iumsc.indiana.edu/index.html
- Structural Classification of Proteins database at Cambridge University: http://scop.mrc-lmb.cam.ac.uk/scop/
The University of California Berkeley mirror site: http://scop.berkeley.edu/ - University of Wisconsin Molecular Modeling Laboratory http://uwmml.pharmacy.wisc.edu/
- BioMagResBank holds structural NMR data for over 1600 proteins and other biomolecules, as well as many useful links.
- Amos' List of Protein 3D structure and associated topics databases. Compiled by Amos Bairoch.
- PDB3D is a 3D molecule rendering Java applet designed specifically for viewing Protein Data Bank (PDB) format molecular structure files within web pages.
Related Topics
Contact information:
John D. Furber
E-mail: johnfurber at aol.com
Telephone: 1-352-271-8711
Gainesville, Florida.
[ John D. Furber home page ]
© 2000 - 2003 by John D. Furber. All rights Reserved.

 The 3-dimensional structures of bio-molecules can give insight into their mechanisms of function, and can suggest strategies for drug design. A number of bio-molecular structures have been solved by two physical techniques: X-ray Scattering from crystals of purified molecules and Nuclear Magnetic Resonance analysis of molecules in solution.
The 3-dimensional structures of bio-molecules can give insight into their mechanisms of function, and can suggest strategies for drug design. A number of bio-molecular structures have been solved by two physical techniques: X-ray Scattering from crystals of purified molecules and Nuclear Magnetic Resonance analysis of molecules in solution.